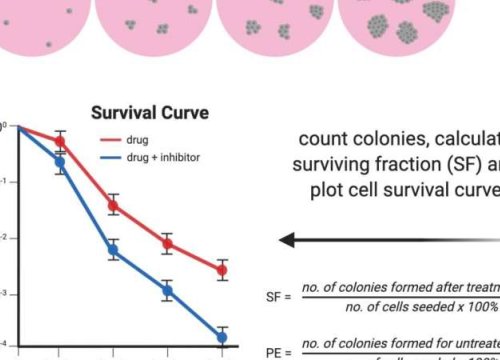
Clonogenic assay or colony formation assay is an in vitro cell survival assay that evaluates the ability of single cells to survive treatment and reproduce to form colonies. Clonogenic assays were initially developed to study the effects of radiation on mammalian cells. More recently, the assays have been widely used to test the efficacy of anti-angiogenic and cytotoxic agents, chemotherapies, and cytokine-based drugs on both neoplastic and normal cells.
With 3D in vitro colony detection assays:
Live-cell imaging for the analysis of colony formation
Live-cell imaging overcomes the drawbacks of traditional clonogenic protocols by enabling scientists to continuously monitor colony growth over long periods of time instead of limiting data acquisition to a single time point.
With live-cell imaging for colony formation:
-
>> Gather more information about the survival of cells following a specific treatment.
-
>> Observe cell division and colony formation in real-time.
-
>> Detect colony formation at a much earlier stage of development, and draw conclusions sooner than when employing a traditional approach.
-
>> Reduce the time and materials needed for your experiment as no endpoint fixation or staining steps are needed.
-
>> Accelerate data analysis and reduce interpretation bias and subjectivity with integrated image analysis software that automatically detects newly formed colonies and evaluates their size and circularity.
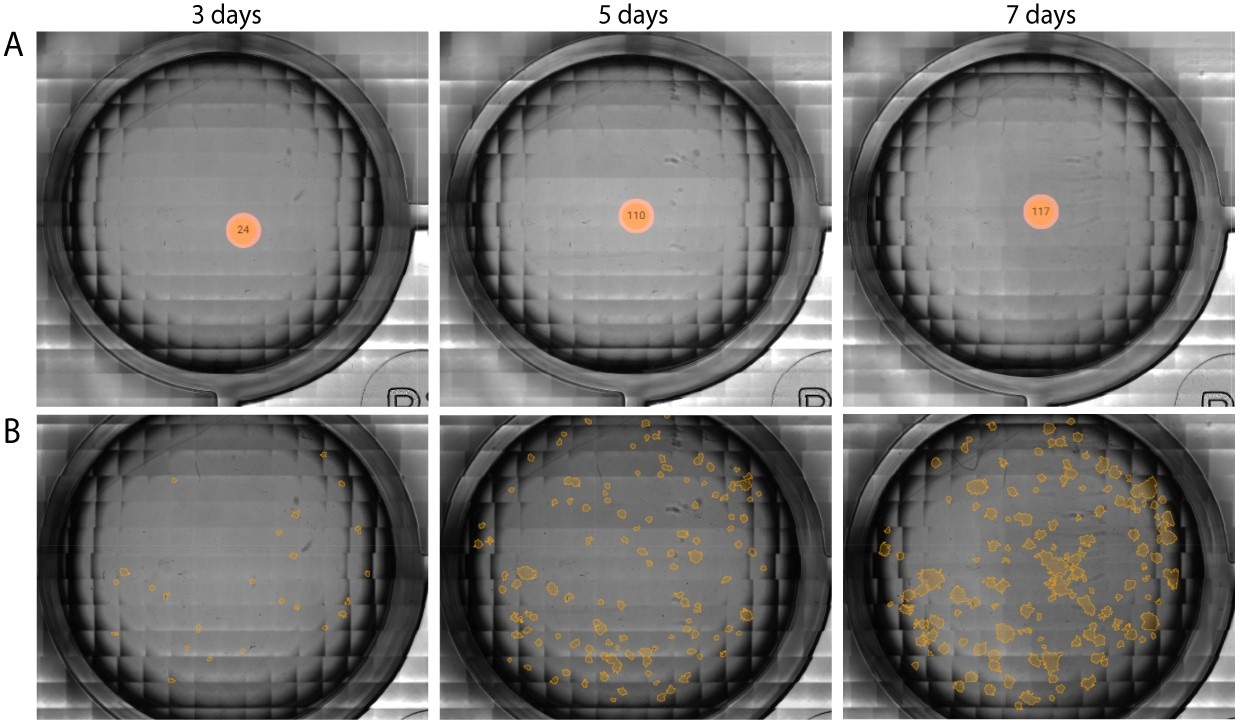
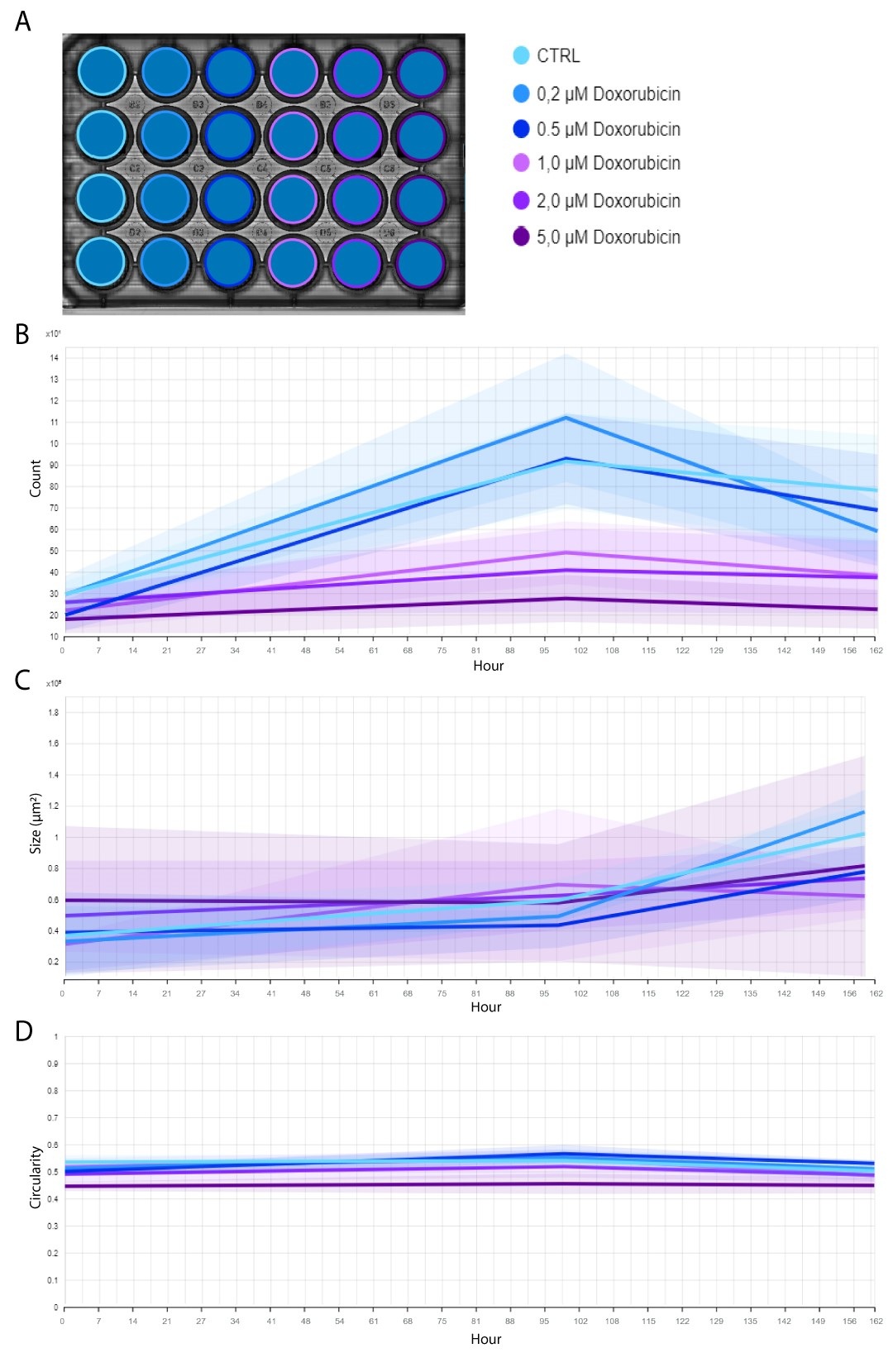
In the example below, CHO-K1 cells, treated with a range of doxorubicin concentrations, were imaged for a total period of 7 days using the Omni. The assay was performed in a 24-well plate that was kept inside a standard CO2-incubator at 37C°.
With the colony detection algorithm, clusters of cells (i.e., colonies) formed in an individual well are automatically quantified (Figure 1A) and highlighted in orange (Figure 1B). This allows users to monitor and quantify cell proliferation and colony formation throughout the entire incubation period (Figure 2A), without missing any critical time points. The algorithm also tracks changes in colony size (Figure 2B) and circularity (Figure 2C).
-
>> Assay your cells in brightfield and fluorescence – From label-free cell monitoring to fluorescence-based assays, add dynamic visual data in any experiment.
-
>> Maintain the optimal culture environment – The Omni and Lux platforms operate within an incubator.
-
>> Track every moment – Automatically capture images as your cells grow in their optimal environment.
-
>> Get started quickly – The live-cell analysis platforms are designed to be simple and flexible. With a host of analysis modules available, it is easy to adapt the systems to meet your needs.