Why measure cell viability?
Cell viability is a measure of the proportion of live cells within a population. The measurement of cell viability plays an essential role in all forms of cell culture. Sometimes, it is the sole purpose of the experiment, for example, in cytotoxicity assays. Alternatively, cell viability can be used to correlate cell behavior to cell number, providing a more accurate picture of cell metabolism. Regardless of the purpose of the experiment, cell viability is extremely important, and there are several methods to assess it accurately.
Cell viability assays – an overview
Cell viability assays usually measure a parameter reflecting the number of live/metabolically active cells in a population. They aim to quantify baseline cellular health and to assess the response to various treatments and stimuli.
A hallmark of dead and dying cells is the breakdown of cell and nuclear membranes. Many viability assays use this characteristic of cell death to distinguish viable from nonviable cells. Cell viability assays can be based on colorimetric, fluorometric, and bioluminescent detection techniques. There is a broad range of in vitro cell viability assays, and there are several decisions to make when selecting the appropriate assays for your needs.
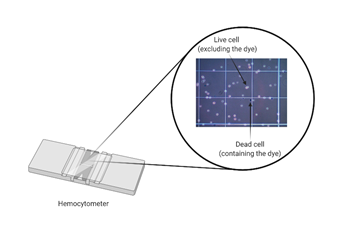
Dye exclusion assays
Dye exclusion methods such as Trypan Blue are commonly used because they are inexpensive and straightforward. These have been developed on the assumption that living cells have intact cell membranes, which will exclude the dye while dying/dead cells will uptake the dye due to impaired membrane integrity, as depicted in Figure 1. Disadvantages include low sensitivity (for example, cells that are still alive but losing cell function will be not accounted for) and the fact that these are endpoint assays, because the dye is extremely toxic for mammalian cells.
Colorimetric assays
Colorimetric assays are based on the measurement of a biochemical marker to evaluate the metabolic activity of the cells. MTT (3-(4,5-dimethylthiazol-2-yl)-2–5-diphenyltetrazolium bromide) assay is one of the most used colorimetric assays to assess cell viability. This assay determines cell viability as a reflection of mitochondrial function. As shown in Figure 2, the MTT assay measures the reduction of MTT by mitochondrial dehydrogenase and the consequent formation of the purple crystal formazan.

ATP-based cell viability assays
Only viable cells can synthesize ATP. Therefore, ATP is an indicator of metabolically active cells, and the number of viable cells in a culture can be measured based on the amount of ATP. ATP cell viability assays are luminometric assays that utilize luciferase to catalyze the formation of light from D-luciferin and ATP.
Fluorescence live-cell imaging for cell viability
All the options that have been described so far are endpoint assays, where the cells need to be killed to obtain a readout. An advantageous alternative can be real-time (live) cell viability assays. Live-cell imaging uses time-lapse microscopy to allow in vivo imaging of cells over a period of time.
The most commonly used technique is live-cell fluorescence microscopy, which allows researchers to follow the localization of different fluorescent proteins in cells1. The first step for this kind of experiment is to introduce a fluorescent reporter in the cell line of interest. Genetically encoded fluorescent protein tags have revolutionized the analysis of intracellular dynamics in the last few decades. State-of-the-art genomic engineering tools such as CRISPR have led to the further development of an even wider variety of fluorescent reporters for tagging proteins.
A common issue during fluorescence live-cell imaging, however, is the occurrence of phototoxicity. Phototoxicity can lead to damage to cellular macromolecules upon light excitation, and this, in turn, can affect sample physiology and potentially lead to cell death2. Limiting it is of the utmost importance to obtain reproducible qualitative and quantitative data on biological samples, although some level of phototoxicity is unavoidable.
Label-free live-cell imaging for cell viability
An alternative to fluorescence-based methods that is gaining popularity is label-free cell imaging. These are non-invasive methods, which use time-lapse microscopy and image analysis algorithms to provide live insight into cellular health. Cellular behavior can be evaluated by comparing e.g. growth rate or migration speed3. One benefit of non-invasive video monitoring is that cell cultures grow as they would in any routine cell culture protocol. Note that this does require that cell cultures are kept in a controlled environment, where temperature, CO2 and O2 levels (in hypoxia chambers) are controlled.
Automated video microscopy holds great potential to assess cell viability and has several advantages over endpoint viability assays or fluorescence-based assays4. Cell death processes are always accompanied by specific morphological features such as volume and shape changes. These can directly be observed and captured using a simple brightfield video system. A time-lapse approach can help to pinpoint not only if, but also when cells start to go into apoptosis.
Tracking of single cells is one application of live imaging that is quite revolutionary. Novel methods use automated tracking software post-imaging. Traditional methods to track cells in populations rely on still images that do not provide an accurate temporal resolution to identify behavioral features in single cells.
------------------------------
Live video monitoring systems such as CytoSMART Lux2 provide an easy and cost-effective way to monitor cell viability in real-time without the need to label the cells.
-----------------------------
Image analysis algorithms that support these live imagining techniques can provide information not only on viability but also on proliferation and cell fate decisions. These are examples of lineage tracing and live population tracking, which can be especially beneficial for stem cell cultures8. Parameters like confluency, area infiltration, and colony growth can be used to generate quantitative data from otherwise qualitative video analysis. Automated analysis of e.g. confluency can provide kinetic information on growth and growth speed.
------------------------------
For researchers looking to perform label-free assays in multi-well plates, we recommend the CytoSMART Omni.
-----------------------------
Other label-free approaches include sophisticated forms of microscopy such as Digital Holographic Microscopy (DHM) and Atomic Force Microscopy (AFM). DHM is able to quantitively and dynamically assess the cellular shape and volume changes with high sensitivity5. AFM has been used as a label-free monitoring and screening technique to assess dying cells recorded in terms of loss of cell adhesion6. In addition, Raman spectroscopy is another versatile tool, which provides a molecular fingerprint of the cells of interest and can be used to assess cell viability in a label-free and high-throughput manner7. A significant advantage of label-free methods over conventional cell viability assays is that they do not require several stages of sample preparation, drastically reducing the time for experiment completion.
Cell viability assays for cell cultures are robust and trusted methods in life sciences. While compound-based assays such as the MTT assay have been around for a long time, these often lack kinetic insight. Novel methods, especially in (label-free) live-cell imaging are presenting themselves as viable alternatives or support to traditional methods.
Related Products
There are currently no products tagged to this resource.