Authors: Marc van Vijven, MSc 1
Oliver Peter, PhD 2
Raphael Lieberherr, MSc 2
1 CytoSMART Technologies B.V., Eindhoven, The Netherlands; 2 Idorsia Pharmaceuticals Ltd, Allschwil, Switzerland
Introduction
We are witnessing a rapid development of techniques to process, analyze and store vast amounts of data. If we can generate more and richer experimental data, now we have the means to turn it into actionable results [1, 2]. In all sectors of life sciences, therefore, there is a trend towards applying large screenings [3]. Clinical laboratories test patient samples for many different biomarkers simultaneously in one diagnostic process, rather than in a sequence of single biomarker tests [4]. Pharmaceutical companies screen for potential drug candidates to treat a disease, before focusing on the most promising compounds [5]. The unprecedented global roll-out of infection testing due to the present Covid-19 pandemic also highlighted the importance of high-throughput assays with large data outputs [6]. The number of samples tested in such screening experiments is too large to be handled manually, so laboratory automation is required. Automation relieves employees from repetitive work, and increases productivity for many tasks, as robots can perform a task faster than humans without needing breaks. Automating processes at any rate increases performance consistency and improves sample and data management [7].
There are several requirements for successful laboratory automation. Firstly, there should be multidirectional communication between all system components, rather than unidirectional control over the devices [8]. In this way, every step in the process is checked by devices confirming the completion of a task. Secondly, automation should ideally cover all steps of a process. Remaining manual interventions can represent bottlenecks for the entire process [9]. These steps do also include the recording, processing, analysis and storage of experimental data. Thirdly, automation requires consistent definitions and protocols, in order to minimize the number of error-prone ‘translation steps’ in the communication between system components [9]. Lastly, there should be flexibility in combining automated modules into a higher-level automation solution [5, 10]. Fixed-purpose, monolithic automation solutions can become useless when the implemented process needs to be changed. Building a system out of multiple modules instead provides flexibility and consequently a long-term benefit.
In this case study we present a total laboratory automation setup including cell culture confluency monitoring at Idorsia Pharmaceuticals Ltd, with the CytoSMART Lux3 FL imaging device, and its integrated image analysis for confluency quantification. All system components communicate using the Standardization in Lab Automation (SiLA 2) communication standard. It provides compatibility to this system, which is assembled from components provided by different manufacturers.
Materials and Methods
Figure 1 displays the automated setup for cell culture confluency monitoring at Idorsia Pharmaceuticals Ltd. The mobile robot collects a well-plate from the incubator and places it on the CytoSMART Lux3 FL (https://cytosmart.com/ products/cytosmart-lux3-fl), which determines the confluency via the integrated image analysis. When the confluency exceeds a trigger value (in this case 70%), the robot moves the wellplate to the automated liquid handler within a biological safety cabinet, which is also remote-controlled, to passage the cells. Otherwise, the well-plate is placed back into the incubator to enable further cell proliferation. The entire workflow is depicted in Figure 2. (Contributing partners to the automated setup: Idorsia Pharmaceuticals Ltd, CytoSMART Technologies, Astech Projects Ltd, UniteLabs AG, i-Gripper GmbH, SiLA).

Fig. 1: Total laboratory automation setup for cell culture confluency monitoring at Idorsia Pharmaceuticals Ltd, with the CytoSMART Lux3 FL for imaging and confluency analysis.
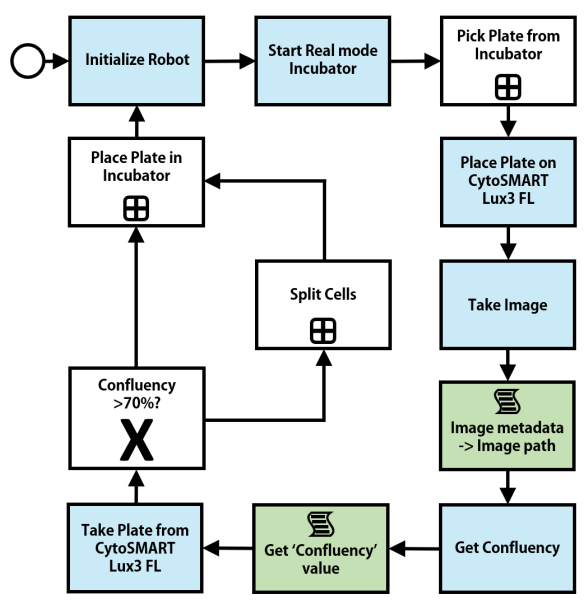
Fig. 2: Workflow and decision-making of automated setup for confluency monitoring at Idorsia Pharmaceuticals Ltd.
Results
The automated setup was used to monitor confluency during expansion for various cell types. Representative images of the SHSY5Y cells (human neuroblastoma cells) and Chinese hamster ovary (CHO) epithelial cells are shown in Figure 3. For both depicted cell types, the setup could easily determine the moment to passage the cells. The SH-SY5Y cells were cultured for approximately 5 days per passage, and the CHO cells grew to sub-confluency every passage in around 2 days.
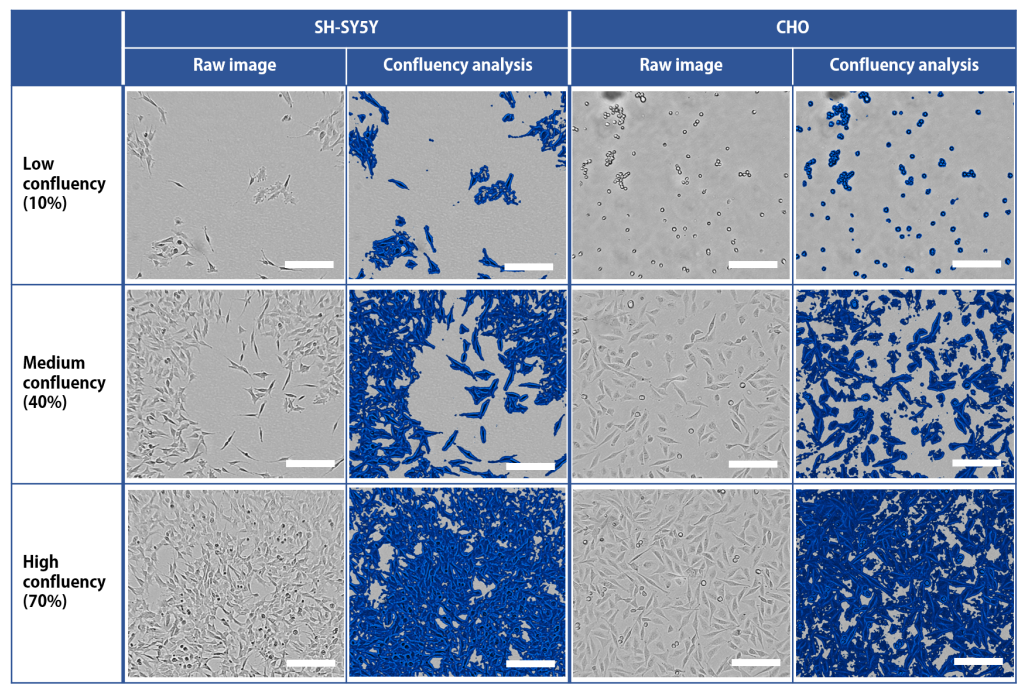
Fig. 3: Examples of confluency monitoring by CytoSMART Lux3 FL in automated setup at Idorsia Pharmaceuticals Ltd. SH-SY5Y cells (human neuroblastoma cells) and CHO cells (Chinese hamster ovary epithelial cells) were monitored up to 70% confluency, which was the moment the cells were automatically passaged. Areas considered to be covered with cells are indicated in blue in the ‘Confluency analysis’ columns. Scale bar = 200 µm.
Discussion
With screening experiments in life science laboratories becoming larger and more common, the need arose to automate these laboratories at least partially. This case study presents a total laboratory automation setup for confluency monitoring at Idorsia Pharmaceuticals Ltd. The CytoSMART Lux3 FL and integrated confluency analysis algorithm play a key role in its decision-making process.
Cell expansion is a continuous and repetitive task in biological laboratories. Automating the process is desirable, freeing employees from this regularly full-time supportive job, and enabling them to perform more creative work. Whereas employees generally are available only during working hours on weekdays – night or weekend shifts being an undesired burden to both the employee and the employer – the automated setup can run non-stop. This obliviates the lag time due to cell preparation on Mondays and mornings, typically seen in manual experiments [11], and thereby contributes to total productivity. Lag time is particularly delaying cell expansion, since cells interrupt their exponential growth phase every time proliferation is disturbed [12]. Besides that, contact inhibition impairs cell proliferation when cells become over-confluent [13], which could happen when they are inspected and passaged too late. The repetitive nature of cell expansion makes it susceptible to inter- and intra-operator variability in confluency estimation and manual task performance, potentially affecting cellular growth. On the other hand, automation thrives when performing repetitive tasks [14], enabling consistent and predictable cell expansion.
The presented automated setup for confluency monitoring at Idorsia Pharmaceuticals Ltd was characterized by the different individual modules, open design of components, and communication standards. The modular system allows easy expansion and adaptation of the setup if required: by adding or adjusting individual modules the entire system can fulfil altered process requirements. The open component design – e.g., the flat stage of the Lux3 FL can handle a wide variety of culture vessels, without requiring specific sample adapters or delicate positioning – provided flexibility in connecting the modules, even though those originated from different manufacturers. Besides this hardware-related connectivity, the SiLA 2 communication standard enabled straightforward connection of and communication between modules on the software side. The results also demonstrated flexibility of the setup regarding the investigated cell type. The system’s ability to determine the confluency was insensitive to the cell morphology and other cell properties, while eliminating user-dependency. Consequently, only minimal adjustments to the imaging and analysis are required when a new cell type is investigated in the existing setup. The time the cells needed to grow to subconfluency matches the reported doubling times for SH-SY5Y cells [15] and CHO cells [16] in literature, which indicates that the cells displayed their regular behavior during expansion in the presented setup.
Conclusion
In this case study, we successfully showed the integration of the CytoSMART Lux3 FL into the automated confluency monitoring system at Idorsia Pharmaceuticals Ltd. The integrated image analysis provided consistent confluency quantification and enabled fast decision-making. The SiLA 2 standard facilitated straightforward communication between all system components.
References
[1] C. Naugler, D. L. Church. "Automation and artificial intelligence in the clinical laboratory", Critical reviews in clinical laboratory sciences, vol. 56, no. 2, pp. 98-110, 2019.
[2] D. Jayasinghe, J. Kimball, S. Choudhary, T. Zhu, C. Pu. "An automated approach to create, store, and analyze large-scale experimental data in clouds", 2013 IEEE 14th International Conference on Information Reuse & Integration (IRI), pp. 357-364, 2013.
[3] N. Blow. "Lab automation: tales along the road to automation", Nature methods, vol. 5, pp. 109-112, 2008.
[4] D. A. Armbruster, D. R. Overcash, J. Reyes. "Clinical chemistry laboratory automation in the 21st century-amat victoria curam (Victory loves careful preparation)", The Clinical Biochemist Reviews, vol. 35, no. 3, pp. 143-153, 2014.
[5] J. Boyd. "Robotic laboratory automation", Science, vol. 295, no. 5554, pp. 517-518, 2002.
[6] J. Hasell, E. Mathieu, D. Beltekian, B. Macdonald, C. Giattino, E. Ortiz-Ospina, M. Roser, H. Ritchie. "A cross-country database of COVID-19 testing", Scientific data, vol. 7, no. 345, 2020.
[7] G. Lippi, G. Da Rin. "Advantages and limitations of total laboratory automation: a personal overview", Clinical Chemistry and Laboratory Medicine (CCLM), vol. 57, no. 6, pp. 802-811, 2019.
[8] S. Zimmermann. "Laboratory Automation in the Microbiology Laboratory: an Ongoing Journey, Not a Tale?", Journal of Clinical Microbiology, vol. 59, no. 3, 2021.
[9] S. Leo, A. Cherkaoui, G. Renzi, J. Schrenzel. "Mini review: Clinical routine microbiology in the era of automation and digital health", Frontiers in Cellular and Infection Microbiology, vol. 10, no. 582028, 2020.
[10] P. P. Bourbeau, N. A. Ledeboer. "Automation in clinical microbiology", Journal of clinical microbiology, vol. 51, no. 6, pp. 1658- 1665, 2013.
[11] M. E. Kempner, R. A. Felder. "A review of cell culture automation", JALA: Journal of the Association for Laboratory Automation, vol. 7, no. 2, pp. 56-62, 2002.
[12] B. Osimani, R. Poellinger. "A protocol for model validation and causal inference from computer simulation", A Critical Reflection on Automated Science, pp. 173-215, 2020.
[13] R. W. Holley, J. A. Kiernan. ""Contact inhibition" of cell division in 3T3 cells", Proceedings of the National Academy of Sciences of the United States of America, vol. 60, pp. 300-304, 1968.
[14] I. Burckhardt. "Laboratory automation in clinical microbiology", Bioengineering, vol. 5, no. 4, pp. 102-114, 2018.
[15] J. Kovalevich, D. Langford. "Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology", Methods in Molecular Biology, vol. 1078, pp. 9–21, 2013.
[16] N. A. Sunstrom, S. Hunt, C. Bailey, M. Baig, M. Sleigh, P. Gray. "Regulated autocrine growth of CHO cells", Cytotechnology, vol. 34, pp. 39-46, 2000.
