Cell proliferation is characterized by cell growth and division, resulting in an exponential increase in total cell number and tissue growth. Various environmental factors (e.g., temperature, pH levels, chemical or infectious agents) can either promote or inhibit cell proliferation, and the study of these factors represents an important area of drug discovery and in vitro cell biology research. The intuitive Cell Confluence Module for the Omni and Lux platforms makes it easy to track and analyze cell proliferation in real time.
With automated cell proliferation analysis:
- >> Monitor and quantify cell growth and proliferation in brightfield and fluorescence
- >> Improve accuracy using whole-well analysis of cell confluence
- >> Assess the effectiveness of biological and chemical agents on cell proliferation
Monitor and quantify cell proliferation in brightfield and fluorescence
Cell proliferation can be used to evaluate normal cell health, to study growth and development of cancer cells, or to measure the effectiveness of cytotoxic agents. The Cell Confluence Module for the Omni and Lux devices, tracks and quantifies cell growth automatically using image analysis.
FIGURE 1: Proliferation of C6 cells over a 5-day period.
Here, C6 glioma cells were imaged for 5 days (120 hours) using the Lux platform. A proliferation curve was automatically generated by the Cell Confluence Module. Over the period of 5 days, the culture progresses to the exponential growth phase, until a plateau is reached at nearly 100% coverage and growth is halted.
Improve accuracy using whole-well analysis of cell confluence
Cells don’t always plate evenly. Omni can image the entire plate calculating confluence across the entire well.
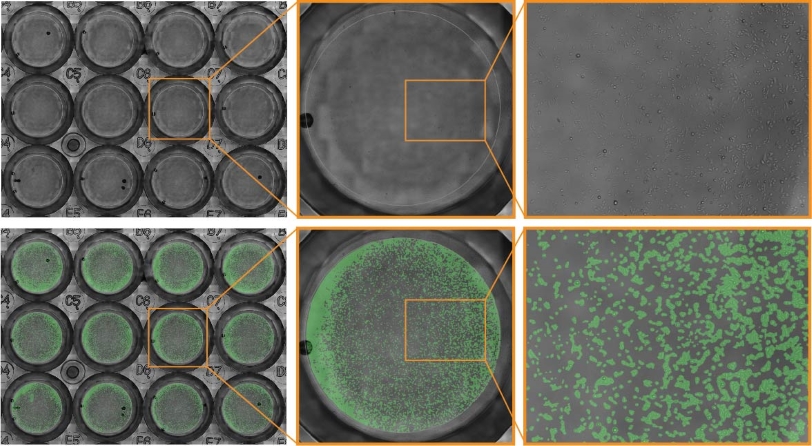
In this example, JnHEK cells were monitored over a 48-hour period. The confluence mask (green overlay) is generated automatically to highlight the position of cells, as detected by the Cell Confluence Module.
Properly time cell passaging using automated cell confluence notifications
The longevity of stem cell cultures requires patience and careful monitoring. Time-lapse imaging can help in keeping track of the culture confluence and any changes that occur in morphology. Especially for iPSCs it is important that the colonies are not over-growing which leads to unstable stem cells. The Omni and Lux platforms can keep track of cell cultures from within the incubator and automated confluency detection monitors cultures closely and unambiguously. The Omni platform is compatible with flasks or plates and can be used to inspect culture homogeneity.
-
>> Assay your cells in brightfield and fluorescence – From label-free cell monitoring to fluorescence-based assays, add dynamic visual data in any experiment.
-
>> Maintain the optimal culture environment – The Omni and Lux platforms operate within an incubator.
-
>> Track every moment – Automatically capture images as your cells grow in their optimal environment.
-
>> Get started quickly – The live-cell analysis platforms are designed to be simple and flexible. With a host of analysis modules available, it is easy to adapt the systems to meet your needs.


